Global Phylogeography and Genomic Epidemiology of Carbapenem-Resistant blaOXA-232–Carrying Klebsiella pneumoniae Sequence Type 15 Lineage
Yuye Wu, Tian Jiang, Xianhong He, Jiayu Shao, Chenghao Wu, Weifang Mao, Huiqiong Jia, Fang He, Yingying Kong, Jianyong Wu, Qingyang Sun, Long Sun, Mohamed S. Draz, Xinyou Xie
, Jun Zhang
, and Zhi Ruan
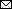
Author affiliations: Zhejiang University School of Medicine Sir Run Run Shaw Hospital, Hangzhou, China (Y. Wu, T. Jiang, X. He, J. Shao, C. Wu, W. Mao, Y. Kong, X. Xie, J. Zhang, Z. Ruan); Wenzhou Medical University Affiliated Wenling Hospital, Taizhou, China (T. Jiang); Third People's Hospital of Xiaoshan District, Hangzhou (X. He, J. Shao); Shaoxing University Affiliated Hospital, Shaoxing, China (W. Mao); Zhejiang University School of Medicine First Affiliated Hospital, Hangzhou (H. Jia); Key Laboratory of Clinical In Vitro Diagnostic Techniques of Zhejiang Province, Hangzhou (H. Jia); Zhejiang Provincial People's Hospital, Hangzhou (F. He); Key Laboratory of Precision Medicine in Diagnosis and Monitoring Research of Zhejiang Province, Hangzhou (Y. Kong, X. Xie, J. Zhang, Z. Ruan); Zhejiang University School of Medicine Fourth Affiliated Hospital, Hangzhou (J. Wu); No. 903 Hospital of PLA Joint Logistic Support Force, Hangzhou (Q. Sun); Hangzhou Women's Hospital, Hangzhou (L. Sun); Case Western Reserve University School of Medicine, Cleveland, Ohio, USA (M.S. Draz); Cleveland Clinic, Cleveland (M.S. Draz)
Main Article
Figure 4

Figure 4. Phylogenetic analysis of global blaOXA-232–carrying carbapenem-resistant Klebsiella pneumoniae sequence type (ST) 15 isolates. A) Time-calibrated maximum clade credibility Bayesian phylogeny based on 330 blaOXA-232–carrying ST15 recombination-filtered core genomes and distribution of antimicrobial resistance genes and virulence genes in the isolates. The cells with colors indicate presence of the gene; blank cells indicate absence. Different colored circles indicate the geographic location and separation time of strains. Blue bars along branches indicate 95% highest posterior probabilities. B) Effective population size of ST15 lineage strains based on the population structure. Shaded areas indicate 95% highest posterior probabilities. Scale bar indicates number of base substitutions per site.
Main Article
Page created: September 05, 2023
Page updated: October 23, 2023
Page reviewed: October 23, 2023
The conclusions, findings, and opinions expressed by authors contributing to this journal do not necessarily reflect the official position of the U.S. Department of Health and Human Services, the Public Health Service, the Centers for Disease Control and Prevention, or the authors' affiliated institutions. Use of trade names is for identification only and does not imply endorsement by any of the groups named above.
