Volume 30, Number 3—March 2024
Dispatch
Highly Pathogenic Avian Influenza A(H5N1) Virus Clade 2.3.4.4b in Domestic Ducks, Indonesia, 2022
Abstract
Highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b viruses were isolated from domestic ducks in South Kalimantan, Indonesia, during April 2022. The viruses were genetically similar to those detected in East Asia during 2021–2022. Molecular surveillance of wild birds is needed to detect potential pandemic threats from avian influenza virus.
The H5N1 subtype of the avian influenza virus A/goose/Guangdong/1/96 (Gs/GD/96) lineage has caused highly pathogenic avian influenza (HPAI) outbreaks in poultry since 1996. In 2008, various novel reassortant viruses were identified in domestic duck and live bird markets (LBMs) in China bearing the genetic backbone of Gs/GD/96 virus clade 2.3.4 hemagglutinin (HA) but different combinations of neuraminidase, such as H5N2, H5N5, H5N6, and H5N8 (1). Clade 2.3.4 continued to evolve into 5th order genetic groups (clades 2.3.4.4a–h); reassortment created different genotypes within those clades (1). H5N8 clade 2.3.4.4 viruses have predominantly spread across many countries in Asia to Europe, Africa, and North America (1,2); repeated outbreaks caused by H5N8 clade 2.3.4.4b viruses were reported during 2016 to mid-2020 (3,4). However, H5N1 clade 2.3.4.4b virus emerged in late 2020, which led to an increase in wild bird and poultry influenza outbreaks worldwide; this virus strain has almost entirely replaced H5N8 clade 2.3.4.4b globally since late 2021 (5). Moreover, the eastward movement of H5N1 clade 2.3.4.4b virus outbreaks from Europe to East Asia since late 2021 suggests that wild birds likely play a role in virus introduction (5,6).
In April 2022, high numbers of poultry deaths were reported from 5 duck farms in Hulu Sungai Utara District, South Kalimantan Province, Indonesia (Appendix Figure 1). Approximately 4,430 of 5,770 (76.8%) ducks of different ages died; younger ducks manifested more severe disease. In July 2023, the deaths of 294 (135 adult and 159 young) of 450 ducks were reported in a Muscovy duck farm in Banjarbaru District of South Kalimantan Province. We collected oropharyngeal swab or tissue samples from ducks in Hulu Sungai Utara in 2022 and Banjarbaru in 2023 for necropsy and hematoxylin/eosin staining; gross and histologic pathology analyses were performed at the Disease Investigation Center Banjarbaru (Appendix). We also collected samples from ducks in LBMs within Banjar District (October 2022), which is located between the Hulu Sungai Utara and Banjarbaru districts where disease was reported (Appendix Figure 1). We sent all influenza A(H5) PCR–positive samples to the Disease Investigation Center Wates in Yogyakarta, where viruses were isolated by using the World Organisation for Animal Health protocol (7). However, viruses could only be isolated from 3 pooled swab samples from the initial cases in April 2022 in Hulu Sungai Utara, 1 tissue sample from the July 2023 case in Banjarbaru, and 1 pooled swab sample from LBMs in Banjar. We characterized the virus isolates antigenically by using hemagglutination inhibition assays and genetically by using whole-genome sequencing on an Illumina sequencing platform (https://www.illumina.com) (Appendix).
We deposited whole-genome sequences of 4 virus isolates into the GISAID database (https://www.gisaid.org) under accession nos. EPI_ISL_17371282 (A/duck/Hulu Sungai Utara/A0522064-06/2022), EPI_ISL_17371283 (A/duck/Hulu Sungai Utara/A0522064-03-04/2022), EPI_ISL_17371284 (A/duck/Hulu Sungai Utara/A0522067-06-07/2022), and EPI_ISL_18438033 (A/Muscovy duck/Banjarbaru/A0523532-9/2023). All 5 identified virus isolates were H5N1 clade 2.3.4.4b viruses, but the virus isolate from LBMs in Banjar District was not included in further analysis or deposited in the GISAID database because of incomplete gene sequences (<50% full-length sequence for each gene segment).
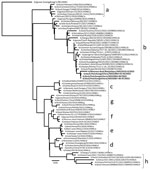
Phylogenetic analysis of the HA gene segment showed that all 4 analyzed viruses clustered with recent HPAI H5 clade 2.3.3.4b viruses from Asia and Europe (Figure). However, they appeared to be more closely related to H5N1 clade 2.3.4.4b viruses from wild birds and poultry from Japan, China, and South Korea isolated during October 2021–February 2022. Phylogenetic trees for the other gene segments (polymerase basic 1, polymerase basic 2, polymerase acidic, nucleoprotein, neuraminidase, matrix protein, and nonstructural segments) also indicated that all 4 viruses were closely related to H5N1 clade 2.3.4.4b from Japan, China, and South Korea (Appendix Figures 2–5). The 3 viruses isolated from the influenza outbreak in April 2022 shared 99.8%–100% nucleotide sequence similarity for each viral segment; however, we observed a lower nucleotide sequence similarity between the viruses from April 2022 and the virus isolated in July 2023 (Table 1), indicating that H5N1 clade 2.3.4.4b continued to mutate resulting in genetic drift. We identified all virus isolates as HPAI on the basis of amino acid sequences within the HA cleavage site (REKRRKR|G); none of those isolates had molecular determinants associated with increased binding affinity or replication efficiency in mammals, including humans (Appendix Table 1) (8,9). A BLAST search (https://www.ncbi.nlm.nih.gov/blast) and pairwise distance analysis indicated all 8 gene segments from viruses isolated during the first outbreak in April 2022 had 98.4%–99.8% nucleic acid sequence identities to H5N1 clade 2.3.4.4b viruses from Japan, China, and South Korea, suggesting a close common ancestor.
The gross and histologic pathology of naturally infected ducks showed multiorgan hemorrhages with prominent lesions in tissues and congestion and focal necrosis in parenchymal cells, often accompanied by inflammatory cell infiltrates (Appendix, Figure 6). Hemagglutination inhibition assays revealed the virus isolates from April 2022 had low reactivity with H5N1 antiserum derived from circulating viruses, including the H5N1 vaccine strains used for poultry (Table 2). Those results suggest that new vaccine candidates antigenically matched to circulating viruses might be needed in Indonesia, if H5N1 clade 2.3.4.4b viruses continue to infect poultry.
Wild migratory birds might play a role in the intercontinental spread of HPAI H5Nx clade 2.3.4.4 viruses (1,10,11). Indonesia is situated within the East Asian Flyway’s island or oceanic routes linking eastern Russia and Japan to the Philippines and eastern Indonesia (12). One stopover site is on the west coast of South Kalimantan, where 23 migratory bird species have been identified and observed (13). Migratory birds often use stopover sites for 1 day to several weeks to rest and refuel (12), providing opportunities for virus transmission through direct or indirect contacts with local wild birds or aquatic poultry within their shared habitats.
During April 2022–July 2023, we conducted molecular surveillance through a network for influenza virus monitoring in Indonesia (14) and did not detect other H5N1 clade 2.3.4.4b outbreaks outside of South Kalimantan. Similar to an earlier virus incursion of H5N1 clade 2.3.2.1c in Java in 2012, which initially also affected ducks (15), we could not determine the exact origin of virus incursion. However, genetic evidence and bird migration patterns suggest that migratory birds contributed to the introduction of H5N1 clade 2.3.4.4b into Indonesia.
We identified HPAI H5N1 clade 2.3.4.4b viruses in ducks in South Kalimantan, Indonesia. The role of migratory birds in virus introduction cannot be ruled out because South Kalimantan is situated within the East Asia Flyway corridor, and the infected farms were connected to marshes that provided opportunity for direct or indirect contacts with migratory birds. Limited wild bird surveillance and genome sequence data for avian influenza viruses impeded our ability to determine further transmission and spread of H5N1 clade 2.3.4.4b in Indonesia. Both epidemiologic studies and molecular surveillance of wild birds are needed to better prepare for pandemic threats caused by continued avian influenza virus evolution in Indonesia and elsewhere.
Dr. Wibawa is a veterinarian at the Disease Investigation Centre Wates, Directorate General of Livestock and Animal Health Services of the Ministry of Agriculture, Indonesia. His research interests focus on molecular diagnostics and epidemiology of influenza viruses in animals.
Acknowledgments
We gratefully acknowledge all data contributors, including the authors and their originating laboratories responsible for obtaining the specimens, and their submitting laboratories for generating the genetic sequence and metadata and sharing via the GISAID Initiative (http://www.gisaid.org), on which this research is based (Appendix Table 2). We also thank the staff at the Disease Investigation Center Banjabaru for conducting field investigation and initial laboratory diagnostics and the staff at the Disease Investigation Center Wates for virus isolation, serology testing, and whole-genome sequencing.
This study was supported by the Directorate General of Livestock and Animal Health Services of the Ministry of Agriculture, Indonesia. Some sequencing reagents and the antigen/antiserum for hemagglutination inhibition assays were funded by the United Nations Food and Agriculture Organization Emergency Centre for Transboundary Animal Diseases, Jakarta, Indonesia and CSIRO-Australian Center for Disease Preparedness, Geelong, Victoria, Australia.
References
- Lee DH, Bertran K, Kwon JH, Swayne DE. Evolution, global spread, and pathogenicity of highly pathogenic avian influenza H5Nx clade 2.3.4.4. J Vet Sci. 2017;18(S1):269–80. DOIPubMedGoogle Scholar
- Kwon JH, Bertran K, Lee DH, Criado MF, Killmaster L, Pantin-Jackwood MJ, et al. Diverse infectivity, transmissibility, and pathobiology of clade 2.3.4.4 H5Nx highly pathogenic avian influenza viruses in chickens. Emerg Microbes Infect. 2023;12:
2218945 . DOIPubMedGoogle Scholar - Yehia N, Naguib MM, Li R, Hagag N, El-Husseiny M, Mosaad Z, et al. Multiple introductions of reassorted highly pathogenic avian influenza viruses (H5N8) clade 2.3.4.4b causing outbreaks in wild birds and poultry in Egypt. Infect Genet Evol. 2018;58:56–65. DOIPubMedGoogle Scholar
- Engelsma M, Heutink R, Harders F, Germeraad EA, Beerens N. Multiple introductions of reassorted highly pathogenic avian influenza H5Nx viruses clade 2.3.4.4b causing outbreaks in wild birds and poultry in the Netherlands, 2020–2021. Microbiol Spectr. 2022;10:
e0249921 . DOIPubMedGoogle Scholar - Xie R, Edwards KM, Wille M, Wei X, Wong SS, Zanin M, et al. The episodic resurgence of highly pathogenic avian influenza H5 virus. Nature. 2023;622:810–7. DOIPubMedGoogle Scholar
- Yang J, Zhang C, Yuan Y, Sun J, Lu L, Sun H, et al. Novel avian influenza virus (H5N1) clade 2.3.4.4b reassortants in migratory birds, China. Emerg Infect Dis. 2023;29:1244–9. DOIPubMedGoogle Scholar
- World Organisation for Animal Health. WOAH terrestrial manual 2021, Chapter 3.3.4. Avian influenza (including infection with high pathogenicity avian influenza viruses) [cited 2023 Jan 2]. https://www.woah.org/fileadmin/Home/eng/Health_standards/tahm/3.03.04_AI.pdf
- Chutinimitkul S, van Riel D, Munster VJ, van den Brand JMA, Rimmelzwaan GF, Kuiken T, et al. In vitro assessment of attachment pattern and replication efficiency of H5N1 influenza A viruses with altered receptor specificity. J Virol. 2010;84:6825–33. DOIPubMedGoogle Scholar
- Suttie A, Deng YM, Greenhill AR, Dussart P, Horwood PF, Karlsson EA. Inventory of molecular markers affecting biological characteristics of avian influenza A viruses. Virus Genes. 2019;55:739–68. DOIPubMedGoogle Scholar
- Caliendo V, Lewis NS, Pohlmann A, Baillie SR, Banyard AC, Beer M, et al. Transatlantic spread of highly pathogenic avian influenza H5N1 by wild birds from Europe to North America in 2021. Sci Rep. 2022;12:11729. DOIPubMedGoogle Scholar
- Zhang G, Li B, Raghwani J, Vrancken B, Jia R, Hill SC, et al. Bidirectional movement of emerging H5N8 avian influenza viruses between Europe and Asia via migratory birds since early 2020. Mol Biol Evol. 2023;40:msad019.
- Yong DL, Heim W, Chowdhury SU, Choi CY, Ktitorov P, Kulikova O, et al. The state of migratory landbirds in the East Asian Flyway: distribution, threats, and conservation needs. Front Ecol Evol. 2021;9:1–22. DOIGoogle Scholar
- Riefani MK, Soendjoto MA. Birds in the west coast of South Kalimantan, Indonesia. Biodiversitas (Surak). 2021;22:278–87.
- Hartaningsih N, Wibawa H, Pudjiatmoko , Rasa FS, Irianingsih SH, Dharmawan R, et al. Surveillance at the molecular level: Developing an integrated network for detecting variation in avian influenza viruses in Indonesia. Prev Vet Med. 2015;120:96–105. DOIPubMedGoogle Scholar
- Dharmayanti NL, Hartawan R, Wibawa H, Balish A, Donis R, Davis CT, et al.; Hardiman. Genetic characterization of clade 2.3.2.1 avian influenza A(H5N1) viruses, Indonesia, 2012. Emerg Infect Dis. 2014;20:671–4. DOIPubMedGoogle Scholar
Figure
Tables
Cite This ArticleOriginal Publication Date: February 13, 2024
Table of Contents – Volume 30, Number 3—March 2024
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Hendra Wibawa, Disease Investigation Center Wates, Jl. Wates KM 27, Wates, Kulon Progo, Yogyakarta 55602, Indonesia
Top