Volume 21, Number 11—November 2015
Dispatch
Fosfomycin Resistance in Escherichia coli, Pennsylvania, USA
Abstract
Fosfomycin resistance in Escherichia coli is rare in the United States. An extended-spectrum β-lactamase–producing E. coli clinical strain identified in Pennsylvania, USA, showed high-level fosfomycin resistance caused by the fosA3 gene. The IncFII plasmid carrying this gene had a structure similar to those found in China, where fosfomycin resistance is commonly described.
Fosfomycin is a phosphonic acid derivative with antibacterial activity against a wide range of gram-negative pathogens and some gram-positive pathogens. It inhibits bacterial cell wall synthesis and is bactericidal against most Escherichia coli strains, as well as many strains of other members of the family Enterobacteriaceae. In the United States, only an oral formulation containing fosfomycin tromethamine is approved for clinical use.
Because of increasing resistance of E. coli strains to other commonly used agents, such as ciprofloxacin and trimethoprim/sulfamethoxazole, fosfomycin has become one of the first-line agents recommended for treatment of uncomplicated urinary tract infection. A recent study of E. coli strains collected at veterans’ hospitals in the United States included ciprofloxacin-resistant and extended-spectrum β-lactamase–producing strains. These strains had 98%–99% susceptibility to fosfomycin (1). However, susceptibility data on fosfomycin are relatively limited overall because this agent is not routinely tested in most clinical microbiology laboratories.
Several fosfomycin resistance mechanisms have been described in E. coli, including reduced permeability, modification of the murA gene target, and modification of fosfomycin (2). In E. coli, the plasmid-mediated fosfomycin resistance gene fosA3, which encodes a glutathione S-transferase, was first identified in a fosfomycin-resistant E. coli strain in Japan (3). This enzyme modifies fosfomycin, thus inactivating the agent and conferring high-level fosfomycin resistance. The fosA3 gene has been reported from only countries in eastern Asia, especially China. We report a case of persistent colonization with a fosA3-carrying E. coli strain in a patient in Pennsylvania, USA.
The patient was a woman with multiple hospitalizations related to sickle cell crises and end-stage kidney disease; she was receiving peritoneal dialysis. She produced minimal amounts of urine and had frequent urinary tract infections due to extended-spectrum β-lactamase–producing E. coli. She did not have history of travel to eastern Asia, from which all reports of fosA3 have so far originated. The first available E. coli strain from this patient, ECRB1, was isolated from a urine sample in 2007 and was reported as a multidrug-resistant strain harboring blaCTX-M-65 and rmtB (4).
She came to a hospital in 2010 because of a power outage in her home. During this brief hospitalization, a culture collected from the peritoneal catheter exit site grew E. coli strain YD472. This strain was found to be highly resistant to fosfomycin, which initiated the present investigation. She had received multiple antimicrobial agents, including cefazolin, ceftriaxone, ciprofloxacin, moxifloxacin, azithromycin, and trimethoprim/sulfamethoxazole, in the prior 5 years, but she had not received fosfomycin according to available medical records.
E. coli YD472 was highly resistant to fosfomycin (MIC >1,024 μg/mL by Etest), which was confirmed by using the agar dilution method with Mueller-Hinton agar and glucose-6-phosphate (25 μg/mL) as an additive, as endorsed by the Clinical and Laboratory Standards Institute (5). Given this high-level resistance, PCR was conducted on YD472 to identify fosA3 and fosC2, which have been reported as acquired fosfomycin resistance genes among recent E. coli strains in Japan, China, and South Korea (3,6,7). A PCR result was positive for fosA3, which was confirmed by sequencing. The fosA3 gene was transferable to E. coli TOP10 by electroporation of the total plasmid extracted from YD472. The E. coli TOP10 transformant strain harboring the fosA3-encoding plasmid pYHCC was resistant to cefotaxime, gentamicin, and fosfomycin.
We then sequenced the entire plasmid in pYHCC by using single-molecule real-time sequencing (Pacific Biosciences, Menlo Park, CA, USA) as described (8). Sequencing in a single cell resulted in a full-length plasmid that could then be circularized and finished (mean coverage 1,325×). The sequence was manually annotated by using RAST (http://rast.nmpdr.org/), ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html), and IS Finder (https://www-is.biotoul.fr/), and comparisons were made by using BLAST analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi). The complete plasmid sequence has been deposited in GenBank under accession no. KR078259.
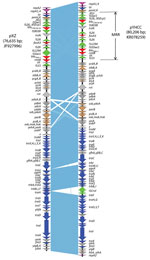
pYHCC is an 80,206-bp circular plasmid with a G + C content of 51.8% and a typical IncFII replicon, which is classified as F2:A–:B– by replicon sequence typing (9). It encodes 121 genes (including hypothetical genes) and harbors a 13-kb multidrug resistance region and a 67-kb R100-like backbone region. The backbone region contains genes for replication, transfer, and maintenance. This backbone of pYHCC contains 24 tra genes and 7 trb genes. The structure of pYHCC is most closely related to that of pXZ, a 77-kb IncFII, fosA3-carrying plasmid reported from E. coli strains isolated from diseased chickens and ducks in China (99% identity with 95% coverage) (Figure 1) (10). It also shares high similarities (99% identity with 84%–87% coverage) with other fosA3-carrying E. coli plasmids reported, including pHN7A8 from a dog in China (11), pHN3A11 from a cat in China, pHK23a from a pig in Hong Kong (12), and pFOS-HK151325 from a human in Hong Kong (13).
A feature that distinguishes pYHCC from pXZ and contributes to the size difference of the plasmids is insertion of ISCroI between trbJ and trbF in pYHCC. Plasmid pYHCC carries 4 antimicrobial resistance genes, all of which are in the multidrug resistance region. These genes are blaCTX-M-65 (conferring cephalosporin resistance), fosA3 (conferring fosfomycin resistance), blaTEM-1 (conferring ampicillin resistance), and rmtB (conferring aminoglycoside resistance). The fosA3 gene and its downstream open reading frame encoding a putative 172-aa protein is flanked by 2 tandem copies of IS26. This arrangement is identical to that in pXZ and pHN7A8, except that pXZ carries blaCTX-M-24 instead of blaCTX-M-65. The blaCTX-M-24 and blaCTX-M-65 gene products differ by only 1 aa (Val for CTX-M-65 and Ala for CTX-M-24 at position 77).
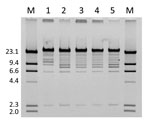
A total of 6 E. coli isolates were collected from the patient during 2007–2010, including the isolates reported here and previously (4). All isolates were obtained from urine samples, except for YD472, which was cultured from the peritoneal catheter exit site. Pulsed-field gel electrophoresis (PFGE) with XbaI as the restriction enzyme showed that all isolates had identical banding patterns. Furthermore, 5 of the 6 isolates, including the first isolate from 2007, were resistant to fosfomycin and positive for fosA3, rmtB, and blaCTX-M-9-group by PCR. E. coli TOP10 transformants harboring fosA3-carrying plasmids were generated from each of these 5 isolates.
The plasmids were then extracted and digested with restriction enzyme EcoRI. The resulting restriction profile was nearly identical across the plasmids except for pYHCC from YD472, which had a shift of a band from ≈8 kb to ≈11 kb (Figure 2). This shift approximated the size difference between pYHCC and pXZ, and further suggested pXZ as the origin of pYHCC.
The first reported clinical E. coli isolate carrying fosA3 was identified in Japan in 2006 (3), but fosA3-carrying E. coli isolates have also been identified in pigs in China as far back as in 2004 (14). Although the patient in Pennsylvania lacked a relevant travel history, the timeline and structural similarities of these fosA3-carrying plasmids make it plausible that this multidrug resistance plasmid originated in E. coli in Asia and moved to the continental United States by human travel or importation of food products. Emergence of E. coli with high-level fosfomycin resistance is a clinically relevant event, especially in the United States, where fosfomycin is increasingly used for empiric treatment of urinary tract infection in the absence of routine drug susceptibility testing.
Dr. Alrowais is a research trainee at the University of Pittsburgh, Pittsburgh, Pennsylvania. Her research interests include emerging mechanisms of antimicrobial resistance in E. coli.
Acknowledgments
This study was supported by the Saudi Arabian Cultural Mission. Y.D. was also supported by research grants from the National Institutes of Health (R01AI104895 and R21AI107302).
Y.D. has served on an advisor board for Shionogi Inc., consulted for Melinta Therapeutics, and received research funding from Merck & Co. for a study unrelated to this study.
References
- Johnson JR, Drawz SM, Porter S, Kuskowski MA. Susceptibility to alternative oral antimicrobial agents in relation to sequence type ST131 status and coresistance phenotype among recent Escherichia coli isolates from U.S. veterans. Antimicrob Agents Chemother. 2013;57:4856–60. DOIPubMedGoogle Scholar
- Castañeda-García A, Blázquez J, Rodríguez-Rojas A. Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics. 2013;2:217–36.
- Wachino J, Yamane K, Suzuki S, Kimura K, Arakawa Y. Prevalence of fosfomycin resistance among CTX-M-producing Escherichia coli clinical isolates in Japan and identification of novel plasmid-mediated fosfomycin-modifying enzymes. Antimicrob Agents Chemother. 2010;54:3061–4. DOIPubMedGoogle Scholar
- Doi Y, Adams-Haduch JM, Paterson DL. Escherichia coli isolate coproducing 16S rRNA nethylase and CTX-M-type extended-spectrum β-lactamase isolated from an outpatient in the United States. Antimicrob Agents Chemother. 2008;52:1204–5. DOIPubMedGoogle Scholar
- Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement M100–S24. Wayne (PA): The Institute; 2014.
- Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3. J Antimicrob Chemother. 2012;67:2843–7. DOIPubMedGoogle Scholar
- Hou J, Huang X, Deng Y, He L, Yang T, Zeng Z, Dissemination of the fosfomycin resistance gene fosA3 with CTX-M β-lactamase genes and rmtB carried on IncFII plasmids among Escherichia coli isolates from pets in China. Antimicrob Agents Chemother. 2012;56:2135–8. DOIPubMedGoogle Scholar
- Li JJ, Lee CS, Sheng JF, Doi Y. Complete sequence of a conjugative IncN plasmid harboring blaKPC-2, blaSHV-12, and qnrS1 from an Escherichia coli sequence type 648 strain. Antimicrob Agents Chemother. 2014;58:6974–7. DOIPubMedGoogle Scholar
- Villa L, Garcia-Fernandez A, Fortini D, Carattoli A. Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J Antimicrob Chemother. 2010;65:2518–29. DOIPubMedGoogle Scholar
- Sun H, Li S, Xie Z, Yang F, Sun Y, Zhu Y, A novel multidrug resistance plasmid isolated from an Escherichia coli strain resistant to aminoglycosides. J Antimicrob Chemother. 2012;67:1635–8. DOIPubMedGoogle Scholar
- He L, Partridge SR, Yang X, Hou J, Deng Y, Yao Q, Complete nucleotide sequence of pHN7A8, an F33:A-:B- type epidemic plasmid carrying blaCTX-M-65, fosA3 and rmtB from China. J Antimicrob Chemother. 2013;68:46–50. DOIPubMedGoogle Scholar
- Ho PL, Chan J, Lo WU, Law PY, Chow KH. Plasmid-mediated fosfomycin resistance in Escherichia coli isolated from pig. Vet Microbiol. 2013;162:964–7. DOIPubMedGoogle Scholar
- Ho PL, Chan J, Lo WU, Lai EL, Cheung YY, Lau TC, Prevalence and molecular epidemiology of plasmid-mediated fosfomycin resistance genes among blood and urinary Escherichia coli isolates. J Med Microbiol. 2013;62:1707–13. DOIPubMedGoogle Scholar
- Hou J, Yang X, Zeng Z, Lv L, Yang T, Lin D, Detection of the plasmid-encoded fosfomycin resistance gene fosA3 in Escherichia coli of food-animal origin. J Antimicrob Chemother. 2013;68:766–70. DOIPubMedGoogle Scholar
Figures
Cite This ArticleTable of Contents – Volume 21, Number 11—November 2015
| EID Search Options |
|---|
|
|
|
|
|
|
Please use the form below to submit correspondence to the authors or contact them at the following address:
Yohei Doi, Division of Infectious Diseases, University of Pittsburgh School of Medicine, S829 Scaife Hall, 3550 Terrace St, Pittsburgh, PA 15261 USA
Top